Determining the causes and evolution of diseases
Pathology consists of examining the cause of the disease, its evolution, its effects on the cells and to make a prognosis. There are three branches of pathology:
- Anatomical pathology is the examination of surgical specimens taken from the body. It includes the analysis of the microscopic aspect of the cells, the immunological markers present in the cells.
- Clinical pathology is the laboratory analysis of body fluids (such as blood, urine or cerebrospinal fluid) and body tissues for the diagnosis of disease.
- Molecular pathology includes analyses based on advanced technologies to determine the molecular characteristics of tumors and to clarify their diagnosis, prognosis and response to targeted therapies. The analysis of nucleic acids (DNA or RNA) extracted from cancerous tissues is part of these analyses which constitute precision medicine.
Diagnosing breast cancer
PathAI , a Boston-based biotech company, participated in the CAMELYON16 challenge, the goal of which was the development of algorithms for automated detection and classification of breast cancer metastases from whole slides of lymph node histology sections.
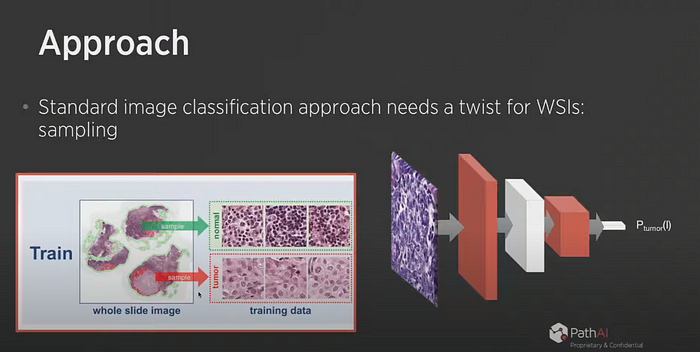
PathAI has developed a model that produces an output heat map on certain parts of the slides “Tumor Probability Map”. A system made available to pathologists who can confirm or not the evaluation of the algorithm by looking at the areas highlighted by the model.

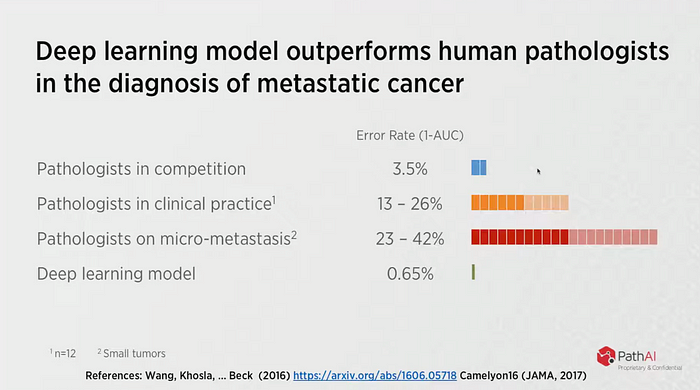
The results of the developed model surpassed the performance of the physicians.
Better selection of candidates for immunotherapy
Analysis of 19 international studies conducted on 11,640 patients with different types of cancers shows that 25% of those treated with immunotherapy showed a “durable response”, compared to only 11% of those who received another family of treatments (chemotherapy or targeted therapy).
Among patients treated with immunotherapy, 30% had an overall survival more than twice as long as the average for all patients, compared to 23% of those treated with other drugs. However, these treatments only work on 20% of patients. Researchers are seeking to better select candidates for immunotherapy.
Tumor cells can evade the immune system’s responses to tumors by expressing a protein called PD-L1 on their surface. PD-L1 works by binding to receptors called PD-1 and B7.1 located on T cells. Binding of PD-L1 to either of these receptors leads to inactivation of T cells.
This binding between PD-L1 and its receptors constitutes an interesting therapeutic target for immuno-oncology. Indeed, blocking the PD-L1 protein can prevent cancer cells from inactivating T lymphocytes through the PD-1 and B7.1 receptors.
Researchers have shown that the more a patient’s tumor expresses PD-L1 protein, the more likely they are to be a good candidate for immunotherapy.
PathAI collected tens of thousands of examples to train a classifier that automatically assesses PD-L1 expression on cancerous tissues. Once trained, the model was applied to two melanoma clinical trials sponsored by Bristol Myers Squibb.

Establishing molecular phenotypes of tumors
The Cancer Genome Atlas Program (TCGA) has generated more than 2.5 petabytes of genomic, epigenomic, transcriptomic and proteomic data. These data are open access and can be used by all members of the research community.
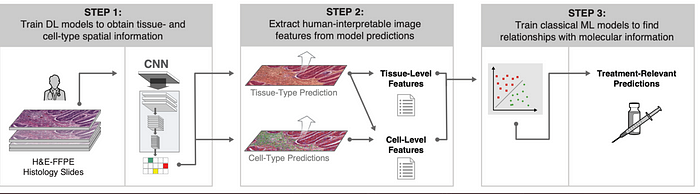
In August 2020, PathAI researchers used 5700 images and 1.6 million annotations made available by TCGA . These images of four cancers (skin melanoma, stomach adenocarcinoma, breast cancer, lung adenocarcinoma) are used to train two convolutional neural networks (CNN)-step 1.

The predictions performed allow the extraction of 607 features called HIFs (human-interpretable image features) that rescind specific and biologically relevant properties of tumor-step 2.
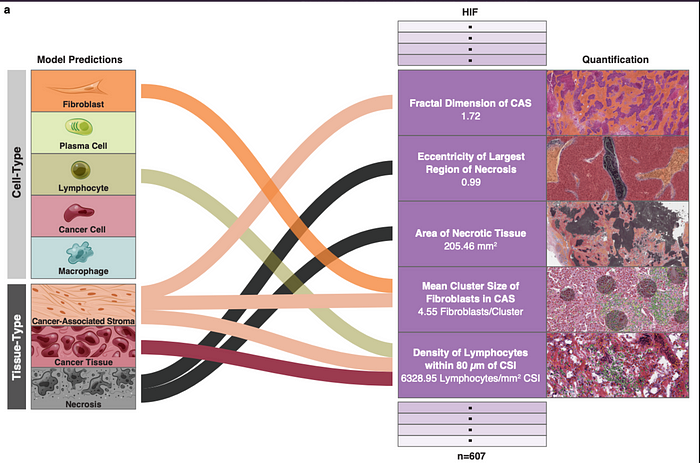
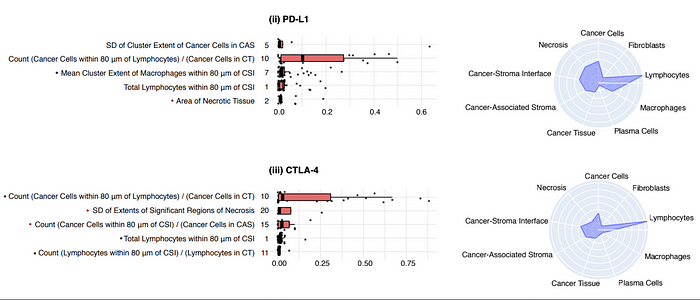
These HIFs by inputting machine learning algorithms allow to predict the presence of PD-1, PD-L1, CTLA-4, TIGIT and HRD molecules in the analyzed tumors of these four cancers. These predictions allow the development of specific treatments and the verification of patients’ eligibility to immunotherapy-step3.
This article is based on these resources
Machine Learning for Pathology — MIT 6.S897 Machine Learning for Healthcare, Spring 2020
More on PathAI
